AI Models and High Performance Server
Brain
- plugin architecture for AI chemistry models,
- Gen3 (Deep Learning), and Gen4 (LLMs)
- including AIMNet2, Microsoft MatterSim and MatterGen
- a base model interface for custom models (enterprise only)
- and a model router for intelligent request routing and load balancing.
- Capable for High Performance like LAMMPS, ORCA …
Pricing, Download and install

Free
Universities, startups
Full model suite (ORB, AIMNet2, MatterSim, MatterGen)
No CUDA, no LLM trainig
Small and Medium Companies
Research groups, corporate R&D teams
Balanced plan for most industrial users.
Contains CUDA and LLM training, and full model suite (ORB, AIMNet2, MatterSim, MatterGen)
$4800 USD*
* per licence, per year
Enterprise
Large pharma, materials companies, HPC centers
- Unlimited model mounts
- Custom plugins
- Dedicated support
For larger dimensions, we welcome the opportunity to partner with you in exploring and optimizing performance. Get in touch to learn more.
Example
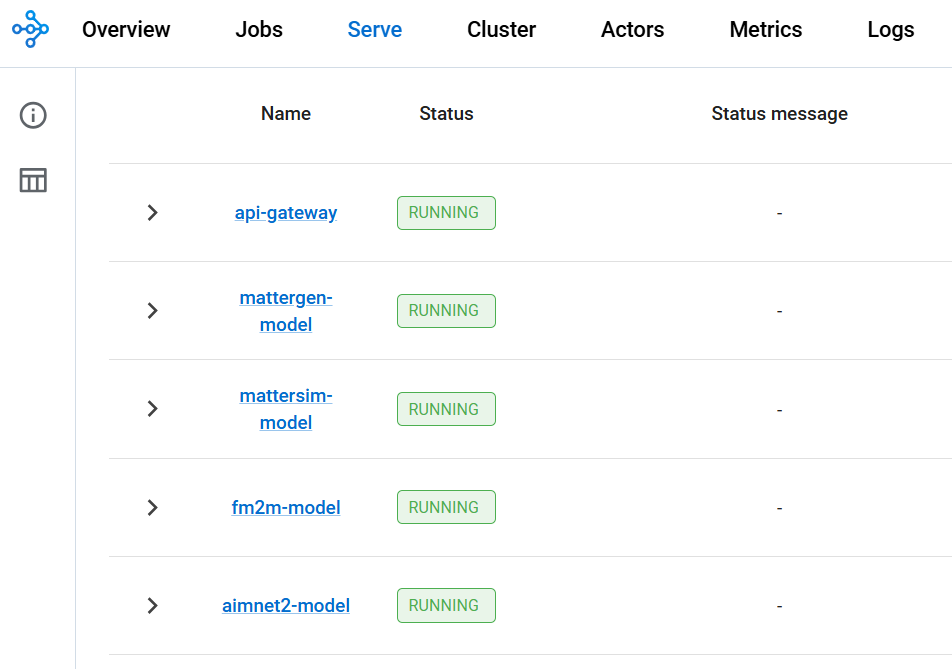
- Models. AIMNet2 (molecular QC), MatterSim (crystal simulation), MatterGen (crystal generation).
- Health/monitoring.
/health,/models,/models/status,/ray/deployments. - Functional tests.
- AIMNet2: H₂, H₂O, CH₄ geometry inquiries; aspirin probe.
- MatterSim: Si diamond cell; graphite layers; perovskite band-structure toy cell.
- MatterGen: Ca₂N; SrTiO₃; Li₂MnO₃; pharmaceutical form of aspirin; MAPbI₃ variants.
- Pipelines: drug-discovery (aspirin); photovoltaic (perovskite).
- Performance. Five-step pseudo-MD trajectory (caffeine) through
/predict. - Acceptance. Presence of
model_used,prediction; graceful degradations flagged as “expected limitation.”
Results
- Auto-routing. Returned “No suitable model found for input type” as an expected limitation; counted as pass.
- Throughput (MD micro-benchmark). 5 steps: mean 18.1 ms (min 16.8, max 20.9 ms) ⇒ ~55 pred s⁻¹ effective.
- AIMNet2 single-shot latencies. H₂ 32.2 ms; H₂O 36.9 ms; CH₄ 39.0 ms; aspirin 37.7 ms.
- Knowledge/synthesis tasks. MatterSim, MatterGen reported 0.0 ms (instrument precision/coarse logging; interpreted as <1 ms or cached).
- Pipelines.
- Drug-discovery (aspirin): QC ✓ (37.7 ms); COX interaction ✓ (<1 ms); crystal-form design ✓ (<1 ms).
- PV perovskite: materials survey ✓ (<1 ms); structure generation ✓ (<1 ms); electronic simulation ✓ (<1 ms).
Representative outcomes
| Domain | Input / Task | Endpoint | Validation | Latency |
|---|---|---|---|---|
| Quantum chemistry | H₂, H₂O, CH₄ geometries | /predict/direct/aimnet2 | model_used=="aimnet2"; prediction present | 32.2–39.0 ms |
| Crystal simulation | Si diamond, graphite | /predict/direct/mattersim | success or expected-limitation handling | <1 ms* |
| Crystal generation | Ca₂N, SrTiO₃, Li₂MnO₃ | /predict/direct/mattergen | prediction present | <1 ms* |
| MD micro-benchmark | Caffeine, 5 steps | /predict (router) | 5/5 responses received | 18.1 ± 1.5 ms |
*Logged as 0.0 ms; treated as <1 ms due to timer resolution/caching.
FAQ
37 HPC engines including ORCA, Psi4, xTB, DFTB+, NWChem, LAMMPS, GROMACS, OpenMM, GULP, ASE, and Quantum ESPRESSO. Plus 7 AI models: ORB, AIMNet2, MatterSim, ANI, MACE, SevenNet, and MatterGen.
No. All engines and AI models run on CPU. No CUDA installation, no GPU drivers, no cloud dependency required.
Geometry optimization, molecular dynamics (NVE/NVT/NPT), electronic structure (DFT, HF, MP2, CCSD(T), CASSCF), thermodynamic properties, IR/Raman/UV-Vis spectroscopy, NMR chemical shifts, and machine learning potentials.
Yes. Brain is accessible via the MCP protocol through Paramus Cloud or the local Chemistry OS. Claude Desktop, VS Code, and Cursor can call Brain engines directly.
Windows for local installation via Chemistry OS, and cloud access from any OS via the MCP server or Paramus Cloud.
