Validated Molecular Descriptor Exchange among AI Agents
SMILES has long been a popular choice for representing chemical structures due to its concise format. However, SMILES strings are inherently linguistic, meaning they rely heavily on textual representation rather than strict structural encoding.
Why SMILES Falls Short: Robust Molecular Descriptors for AI
This linguistic characteristic makes SMILES vulnerable to subtle, yet critical, alterations by Large Language Models (LLMs), potentially causing loss of structural fidelity. Maintaining molecular descriptor fidelity thus requires robust, non-ambiguous representations beyond SMILES, like Molfile V3000, InChI, or structured JSON formats.
Round-Trip Fidelity: Safeguarding Chemical Data in AI Pipelines
Round-trip fidelity, the capability to maintain exact molecular descriptor integrity when transferring data between computational agents, is essential in cheminformatics. Achieving this ensures that a molecule retains its precise structure throughout interactions with LLM-driven workflows, preventing unintended changes that could compromise research accuracy.
Descriptor Reliability in AI Chemistry: The PARAMUS Approach
To address this, PARAMUS is actively developing an architecture that secures high-quality molecular descriptors across interactions involving LLMs and AI agents. By implementing a centralized molecule registry utilizing redundant formats — such as canonical SMILES, InChI, and explicitly defined Molfile formats — PARAMUS ensures reliable round-trip descriptor fidelity. The system additionally employs strict validation steps at every data handover point, ensuring precise molecular information retention.
High-Fidelity Molecular Data Handling in LLM-Orchestrated Pipelines
A centralized molecular registry storing redundant descriptor formats ensures precise round-trip fidelity between AI agents and LLMs, preserving molecular integrity in cheminformatics workflows.
All agent interactions reference molecule by ID, avoiding direct descriptor manipulation.
- User submits V3000 Molfile to the LLM orchestrator.
- LLM orchestrator delegates molecule registration to cheminformatics tools (RDKit/Open Babel).
- Centralized database (RelDB) stores molecule in multiple canonical forms (Molfile V3000, SMILES, InChI, JSON).
- All agent interactions reference molecule by ID, avoiding direct descriptor manipulation.
- Descriptor Fidelity Validator ensures integrity after each transformation.
- Round-trip verification confirms descriptor preservation before returning results to the user.
Integrity Matters: Molecular Descriptor Preservation in AI Agents
This structured approach significantly enhances the reliability of AI-driven cheminformatics pipelines, facilitating accurate and reproducible chemical analyses. As computational chemistry increasingly integrates with advanced AI, adopting such rigorous data handling methods becomes imperative.
Philosophically, the proposed architecture highlights the tension between symbolic and subsymbolic representation in computational chemistry involving LLMs. Symbolic representations, like structured molecular descriptors (e.g., Molfile V3000, InChI), offer:
- explicit,
- discrete, and
- semantically clear
encoding of molecular structures.
Bridging Symbolic Accuracy and Subsymbolic Flexibility: Integrating Molfiles and SMILES
Conversely, subsymbolic approaches (such as neural embeddings used by LLMs) encode meaning implicitly, relying on statistical patterns rather than defined rules. This architectural approach reconciles these opposing paradigms by strictly separating symbolic descriptor management (handled explicitly in a robust database structure) from subsymbolic inference (handled by LLMs).
Combining symbolic precision (Molfile V3000) with the linguistic advantages of SMILES for LLMs can be effectively achieved by storing both descriptors side-by-side, linked via a unique identifier. Specifically:
- Molfile V3000 (Symbolic):
Ensures exact chemical structure fidelity, preserving coordinates, stereochemistry, and explicit structural relationships. - Canonical SMILES (Subsymbolic-Friendly):
Provides LLM-friendly linguistic representation, facilitating semantic reasoning, chemical similarity judgments, and text-based pattern recognition.
In practice, the LLM references molecules via a unique ID, retrieving the SMILES representation for semantic and chemical context inference, while the exact symbolic structure (Molfile) is reserved for precise cheminformatic computations. Thus, both symbolic (exact chemical accuracy) and subsymbolic (LLM linguistic reasoning) paradigms coexist, maximizing their respective strengths without compromising data fidelity.
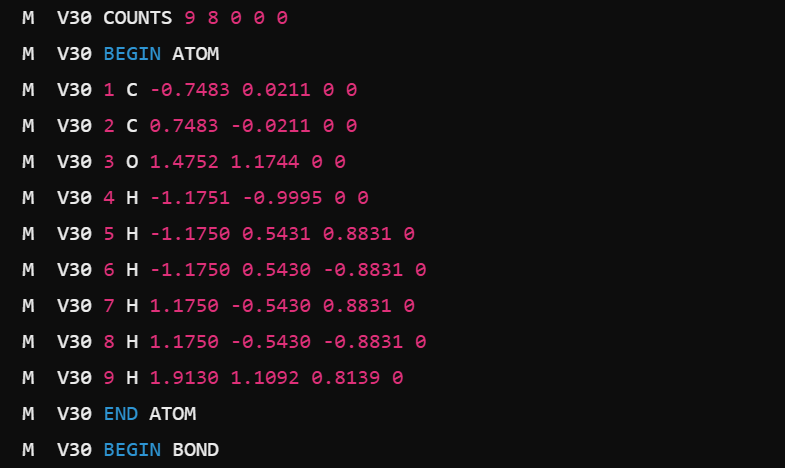
Molfile V3000 Example
PARAMUS is currently implementing this architecture and plans to roll it out with version 2 in autumn 2025.
PARAMUS’s commitment to robust descriptor management positions it at the forefront of reliable cheminformatic integration with emerging AI technologies.
